 |
Home |
| Genome-Phenome Analyzer |
- Clinical findings and lab tests input by the user, providing phenotype information to be compared to the phenome.
- Annotated genomic variants, providing genotype information to be compared to the genome.
The approach is distinctive in being hypothesis-independent in not requiring assumptions about mode of inheritance, number of genes involved, or which clinical findings are most relevant. In some clinical situations it is possible to construct a short list of genes that are relevant (e.g., for centronuclear myopathy) but it many other situations the clinical picture is less pathognomonic and a more robust clinical analysis is needed.
This is enabled by a curated, evidence-based, computational database of diseases and findings is combined with a Bayesian approach (no evoking strengths) to enable it to reason from pertinent positives and negatives. This approach allows use of timing of onset and disappearance information about individual findings, along with family history, and taking cost into consideration in test recommendations. More details about the approach and the efficacy are in a study published in the Journal of Child Neurology (free PubMed Central and PDF versions).
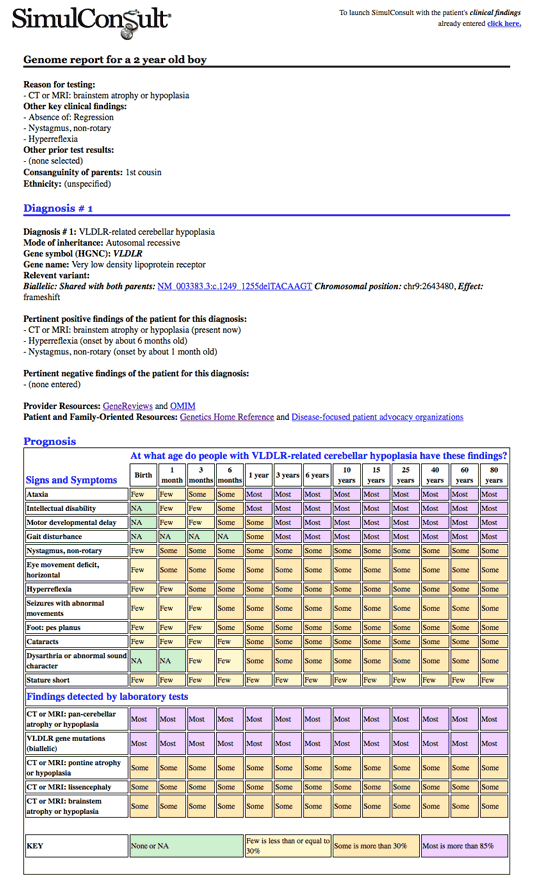
The Genome-Phenome Analyzer now outputs incidental findings with options for gene lists and individuals to report. New features allow the user to quickly select the reasons for testing and up to 2 diagnoses and associated genes and variants, as well as the incidental findings and any prioritized gene discovery lists. These can be exported to a lab's reporting platform using a standard API. The software automatically includes the prognosis table to answer the clinician and patient question "what should I expect?" and assigns the patient's findings to the relevant diagnosis.
Learn about new features here.
